BONAS
BO - https://brunch.co.kr/@tristanmhhd/19
https://wooono.tistory.com/102
[Abstract]
estimation strategy:
- one-shot < sample-based: more reliable
⇒ BONAS: accelerate, keeps reliability
[overview of BONAS]
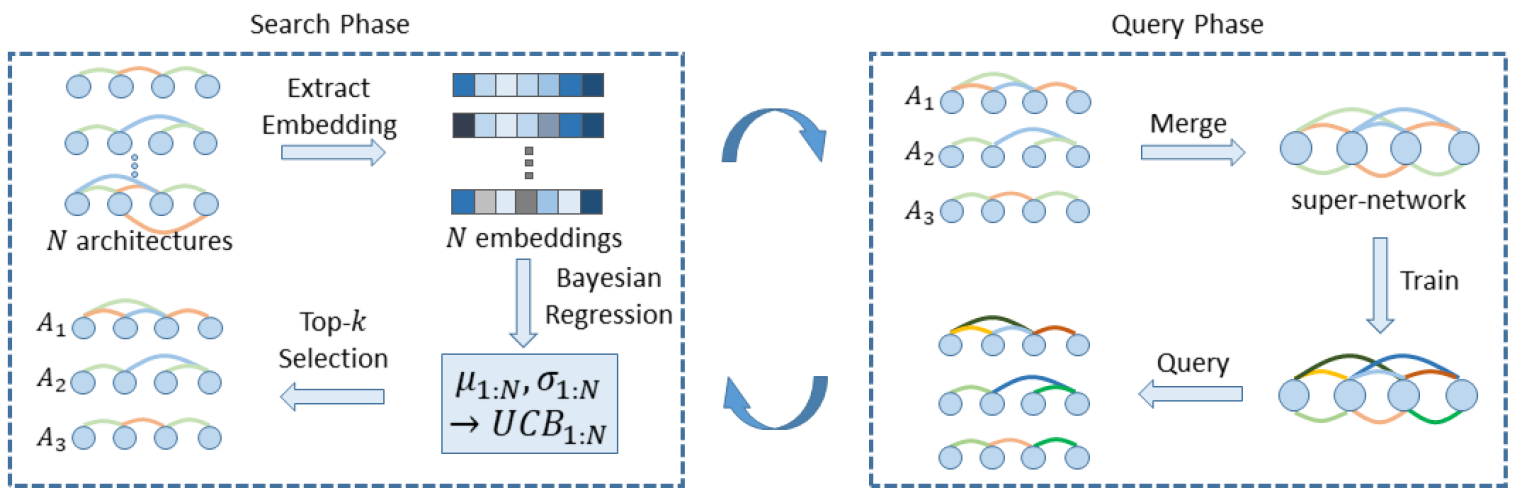
- searching for competitive architectures is difficult ⇒ BO handles this issue by explicitly considering exploitation and exploration
- In BO, a commonly used surrogate model is GP
- prob 1: cubically with the number of samples)
- prob 2: requires well-designed kernel (open issue)
- one-shot methods: weight-sharing is performed on the whole space of sub-networks. → not good idea
⇒ costly in NAS
- query phase: traditional approach of fully training neural architectures is costly
⇒ solution : BONAS - sample-based NAS algorithm combined with weight-sharing
- search phase: use GCN to produce embeddings for NA (avoid to define GP’s kernel function, replace GP → novel Bayesian sigmoid regressor)
- query phase: construct a super-network from candidate architectures → train by uniform sampling → candidates are queried simultaneously based on the learned weight of the super-network
⇒ BONAS outperforms SOTA methods
[algorithm]
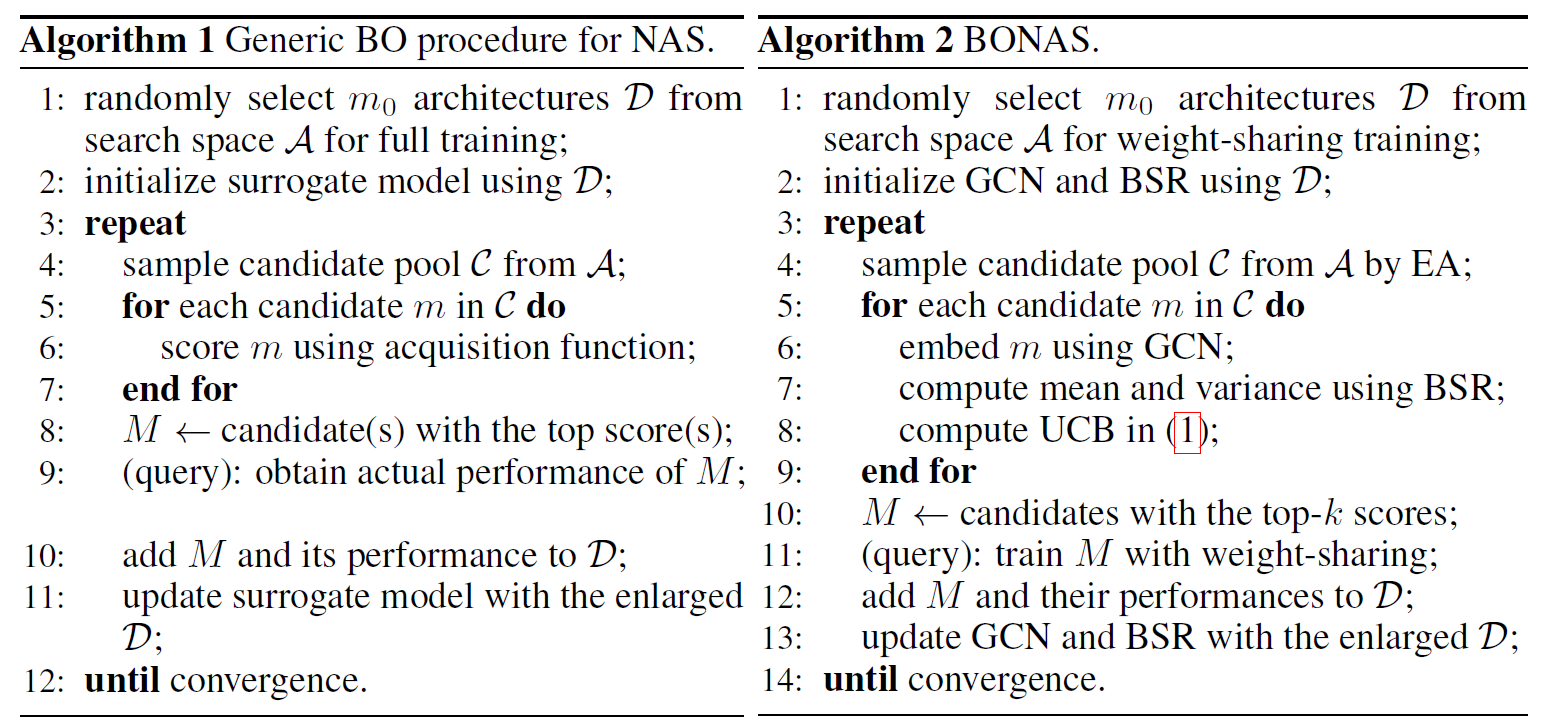
Q: three main points of BO in Algorithm 1
- represent architecture
- find good candidates with surrogate model (high acuisition score)
- query the selected candidates
⇒ rather full training in each query (cost), adopt weight-sharing paradigm to query a batch of promising architecture together (비슷한 것끼리 weight sharing?)
⇒ design a surrogate model combining GCN embedding extractor and Bayesian sigmoid regressor
[BO: surrogate model, acquisition function]
- surrogate model: GCN embedding extractor + BSR → get mean and variance
- acquisition function: UCB
- GCN embedding extractor
- NAS-Bench-101: stacking multiple repeated cells
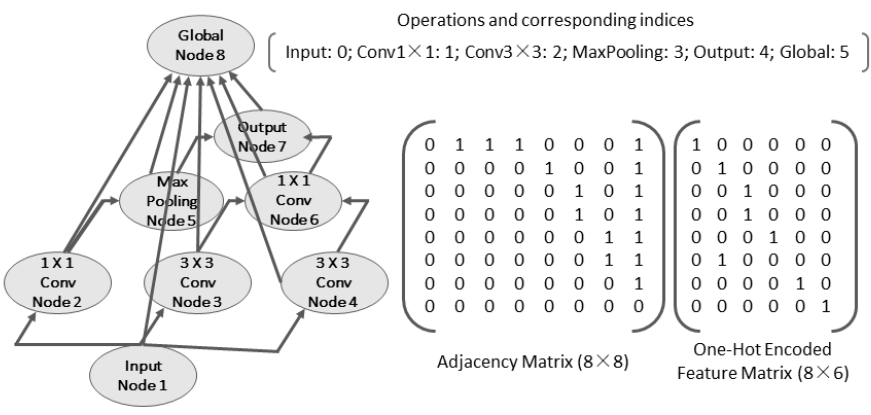
- MLP or LSTM used to encode networks, but GCN is better
- global node: to get the embedding of the whole graph
In experiment,
- use single-hidden-layer network with sigmoid function, prediction: [0,1]
- regressor is trained by minimizing square loss
⇒ We get embedding of a graph(cell) by GCN!
- Bayesian Sigmoid Regression(BSR)
- We’re interested in UCB (Upper Confidence Bound) of BO.
- compute the mean and variance of architecure’s performance
-
Since final layer of GCN predictor contains a sigmoid, instead of fitting true performance t, estimate the value before sigmoid. y=logit(t) ⇒ nonlinear regression → linear regression
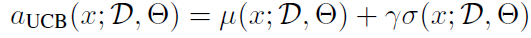
-
GCN: squared loss → BSR: exponentially weighted loss (emphasis on model with higher accuracies)
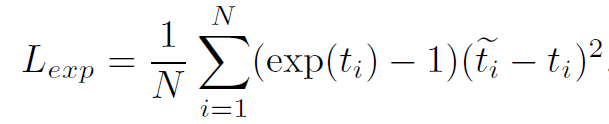
-
For candidate network (A, X), BO has to evaluate its acquisition score. For UCB, the mean of logit(t) and variance of logit(t) are:
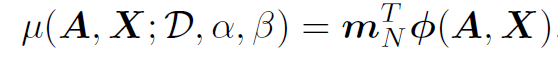
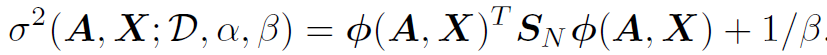
- Then, we can convert those to predict mean and variace of “t” by
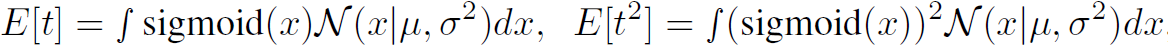
- Then, we can approximate E[t] and Var[t] by using $\Phi(\lambda x)$ for some $\lambda$ and $\Phi$.
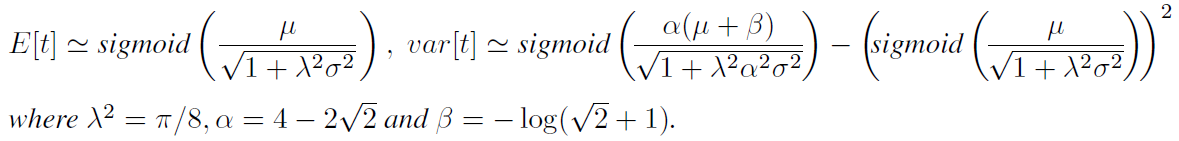
- Then we can put E[t] and Var[t] into UCB score and use this to select the next architecture sample from the pool.
[Efficient Estimation]
- NAS: select architecture in each iteration for full training ⇒ expensive
- BONAS: select in each BO iteration a batch of k architectures $\set{(A_i, X_i)}_{i=1}^k$ with the top-k UCB scores → train them together as a super-network by weight sharing
{advantage of weight sharing of super-network}
- feasible to train each architecture fairly
- super-network is promising candidate because of their high UCB scores
{step}
-
construct the super-network using logical OR operation
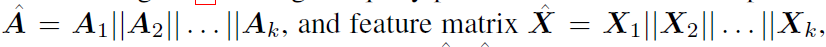
-
super-network is then trained by uniformly sampling from the architectures
- one sub-network is randomly sampled from the super-network
- only the corresponding (forward, backward propagtion) paths are activited)
- evaluate each sub-network by only forwarding data along the corresponding paths in the super-network
[Summary of Algorithm]
- Get embedding of each graph by GCN embedding extractor
- For each search iteration, a pool C of candidates are sampled from A by EA (each candidate’s embeding is obtained in 1.). For each candidate, compute UCB by BSR
- Candidates with the top-k UCB scores are selected(super-network) and queried using weight sharing
- evaluteed model and their performance values are added to D
- GCN predictor and BSR are updated using the enlarged D
[Experiments]
-
dateset
[convlutional achitectures]
- NAS-Bench-101: 423K
- NAS-Bench-201: 15K architectures, using different search space
[Others]
- LSTM-12K (containing 12K LSTM models trained on the Penn TreeBank dataset)
-
comparison: compare with existing MLP and LSTM preditors, and the meta NN - our GCN has four hidden layers with 64 units each
- square loss
- Adam optimizer
- 0.001 learning rate
- mini-batch of size 128
- 8.5:1.5:0.5
[closed domain search]
- compare with SOTA sample-based NAS baselines
- random search, regularized evolution, NASBOT, NAO, LaNAS, BANANAS
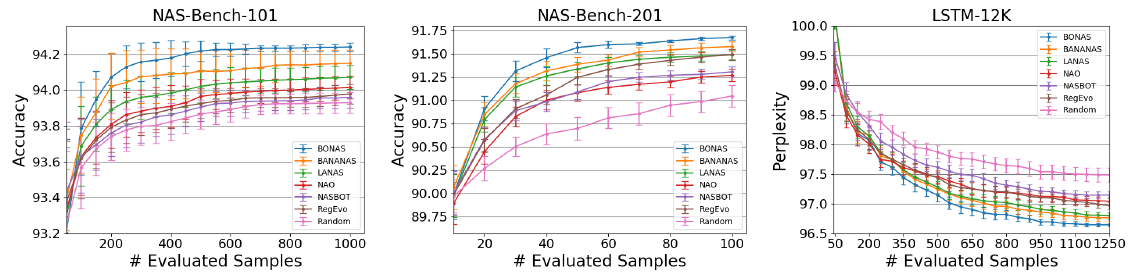
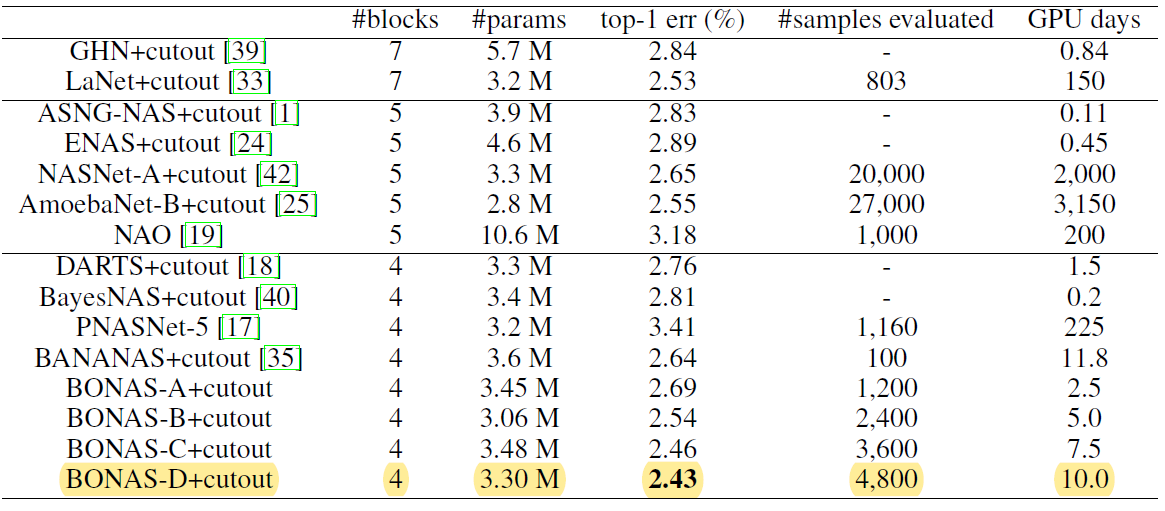
[open domain search]
- perforam NAS on NASNet search space using CIFAR-10 data set
- allow 4 blocks inside a cell
- k=100 models are merged to a super-networ, and trained for 100 epochs
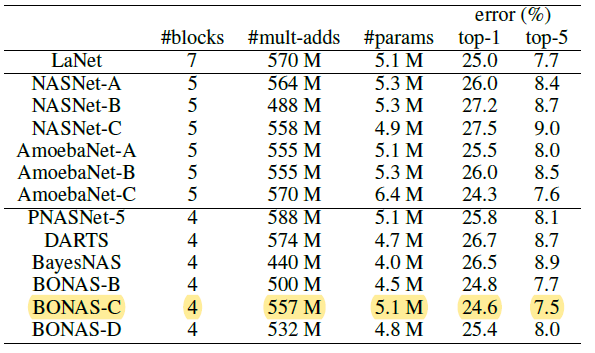
- compare with SOTA
- sample-based NAS algorithms: NASNet, AmoebaNet, PNASNet, NAO, LaNet, BANANAS
- one-shot: ENAS, DARTS, BayesNAS, ASNG-NAS
- A, B, C, D: different numbers of evaluated samples
[transfer learning]
- transfer the architectures learned from CIFAR-10 to ImageNet.
-
Use BONAS-B/C/D
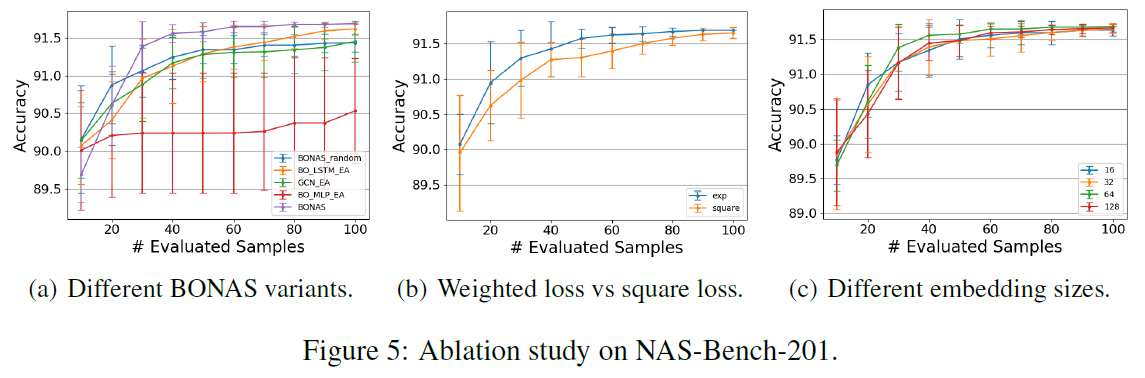
[Ablation Study]
- study BONSA variants (5-a)
- BONAS_random: EA sampling → random sapling
- BO_LSTM_EA: GCN predictor → LSTM
- BO_MLP_EA: GCN predictor → MLP
- GCN_EA: removes Bayesian sigmoid regression and uses GCN output directly
⇒ BONAS outperforms others
- proposed weight loss vs traditional square loss (5-b)
- verify the robustness of BONAS model (5-c)
[closed domain search]
- compare with SOTA sample-based NAS baselines
- random search, regularized evolution, NASBOT, NAO, LaNAS, BANANAS

[open domain search]
- perforam NAS on NASNet search space using CIFAR-10 data set
- allow 4 blocks inside a cell
- k=100 models are merged to a super-networ, and trained for 100 epochs
- compare with SOTA
- sample-based NAS algorithms: NASNet, AmoebaNet, PNASNet, NAO, LaNet, BANANAS
- one-shot: ENAS, DARTS, BayesNAS, ASNG-NAS
- A, B, C, D: different numbers of evaluated samples

[transfer learning]
- transfer the architectures learned from CIFAR-10 to ImageNet.
-
Use BONAS-B/C/D

[Ablation Study]
- study BONSA variants (5-a)
- BONAS_random: EA sampling → random sapling
- BO_LSTM_EA: GCN predictor → LSTM
- BO_MLP_EA: GCN predictor → MLP
- GCN_EA: removes Bayesian sigmoid regression and uses GCN output directly
⇒ BONAS outperforms others
- proposed weight loss vs traditional square loss (5-b)
- verify the robustness of BONAS model (5-c)
