[abstract]
- GNN에서도 pre-training이 효과적인가
- pre-training GNN에 대한 새로운 self-supervised method 개발
- individual node 단위에서 학습
- entire graph에서 학습
- 분류 문제에서 multiple graph를 pre-train함
- naive approach는 성능 개선을 보여주지 못함
- 우리의 approach는 성능 개선을 보여줌
- Introduction
- pre-training is useful
- task-specific labeled data can be extremely scarse (chemistry, biology)
- real world graph data에는 가끔씩 out-of-distribution samples가 있음 - application to graph is hard
- requires domain expertise to select examples and target labels (correlated with the downstream task of interest)
- otw, harm generalization → negative transfer - contributions
- conduct investigation of strategies
- develop effective pre-training strategy for GNNs
- capture domain-specific knowledge about nodes → graph-level knowledge
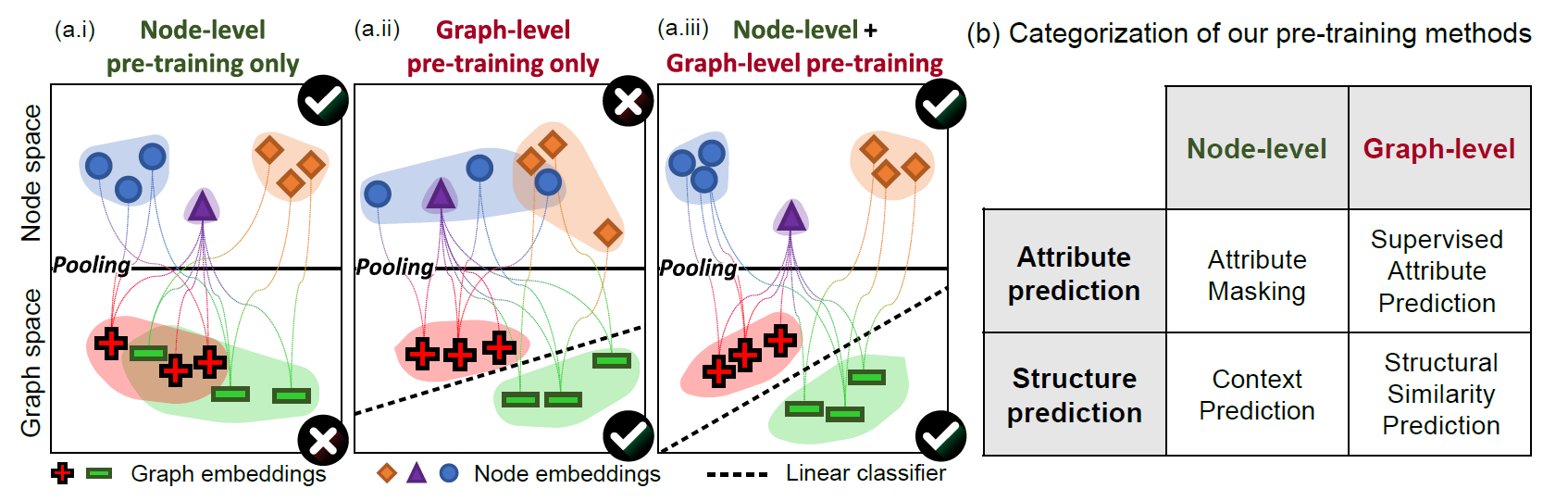
- a.i: node-level: node끼리 cluster
- a.ii: graph-level: graph끼리 cluster
-
preliminaries of GNNs
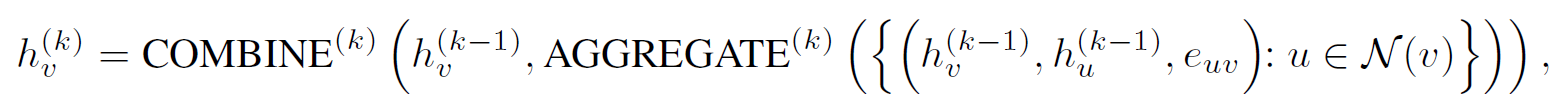
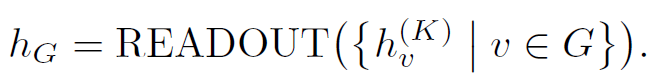
- Strategies for pre-training GNNs
3.1 Node-level pre-training
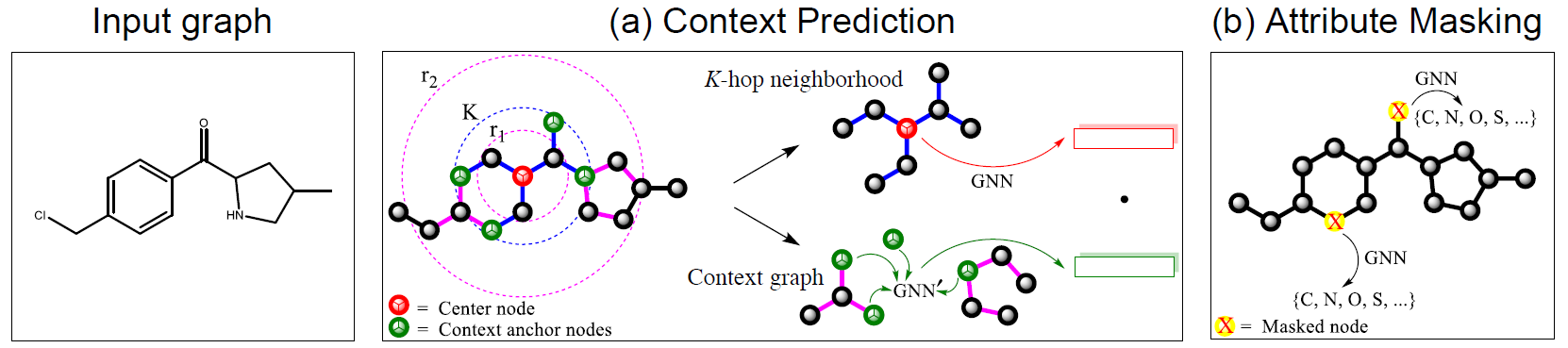
- use unlabled data to capture domain-specific knowledge
- context prediction
-
structure
(1) k-hop neighbors
(2) context graph: subgraphs between r1-hops and r2-hops (r1<K)
⇒ with b and c, we know how neighborhood and context graphs are connected with each other
- encoding method (context → fixed vector)
- context graphs → fixed-length vectors
- use auxiliary GNN (context GNN)
- learning via negative sampling
-
목표: neighborhood와 context graph의 node가 같은 node인지
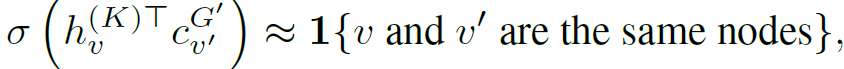
- v’=v, G’=G : positive neighborhood-positive pair
- randomly sample v’ from random G’: negative neighborhood-context pair
- sampling ratio: one nagative: one positive
-
-
- attribute masking
- input node/edge attributes are randomly masked ⇒ predict
- example
- molecular graph: node attributes (atom type)
- PPI: edge attributes (interactions)
- context prediction
3.2 Graph-Level pre-training
- method : making predition about
- domain-specific attributes
- properties of molecules (supervised tasks)
- naive performing may be fail → may be unrelated to the downstream task
- regularize GNN at the level of individual nodes via node-level pre-training methods
- graph structure
- task: graph edit distance, structure similarity
- graph distance values → difficult problem… ⇒ left this future work
- domain-specific attributes
3.3 Overview
[pre-training]
- step1: node-level self-supervised pre-training
- step2: graph-level multi-task supervised pre-training
[fine-tuning]
- linear classifiers to predict graph labels
- experiments
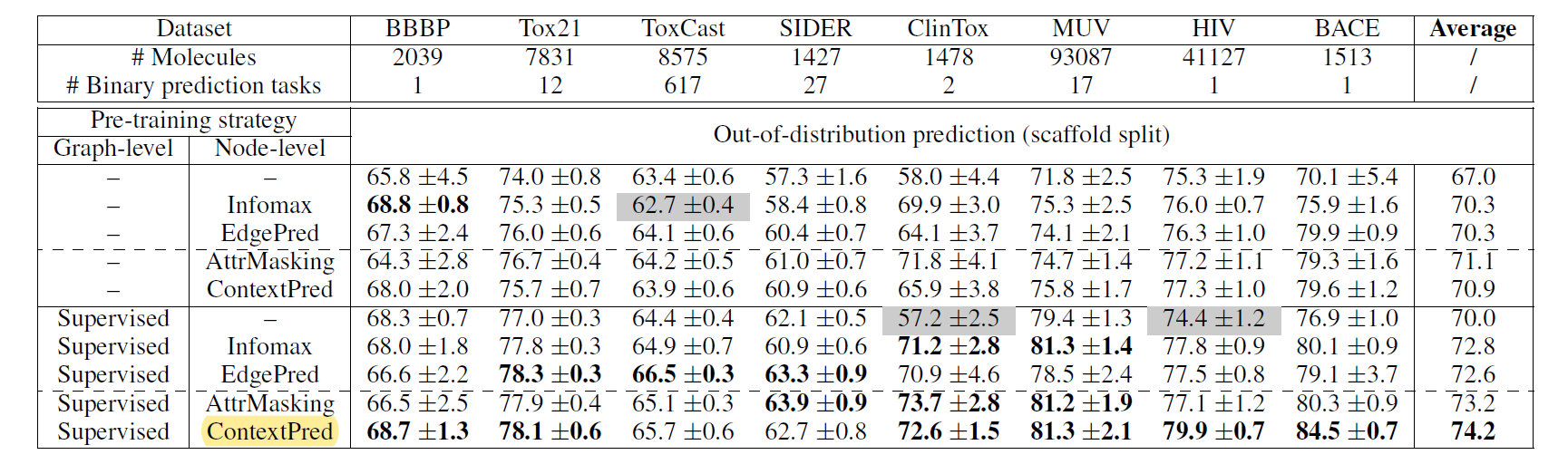
5.1 datasets
- data splitting
- molecules: scaffold split → solve out-of-distribution generalization
- biology: species split
[result]
[apply to graph NAS]
- node level: attribute masking
- graph level: graph edit distance
- fine tuning