[Abstract]
- introduce a novel graph generative model: Computation Graph Transformer (CGT)
- generates effective benchmark graphs on which GNNs show similar task performance as on the source graphs
- scales to procecss large-scale graphs
- incorporates off-the-shelf privacy modules to guarantee end-user privacy of the generated graph
[1. Introduction]
- graph data: privacy-restricted
- want to overcome the limited access to real-world graph datasets
- goal: generate synthetic graphs that follow its distribution in terms of graph structure, node attributes, and labels (substitues for the original graph)
- problem statement: for a given G=(A, X, Y) (A: adjacency, X: node attributes, Y: node label), generate G’=(A’, X’, Y’) satisfying
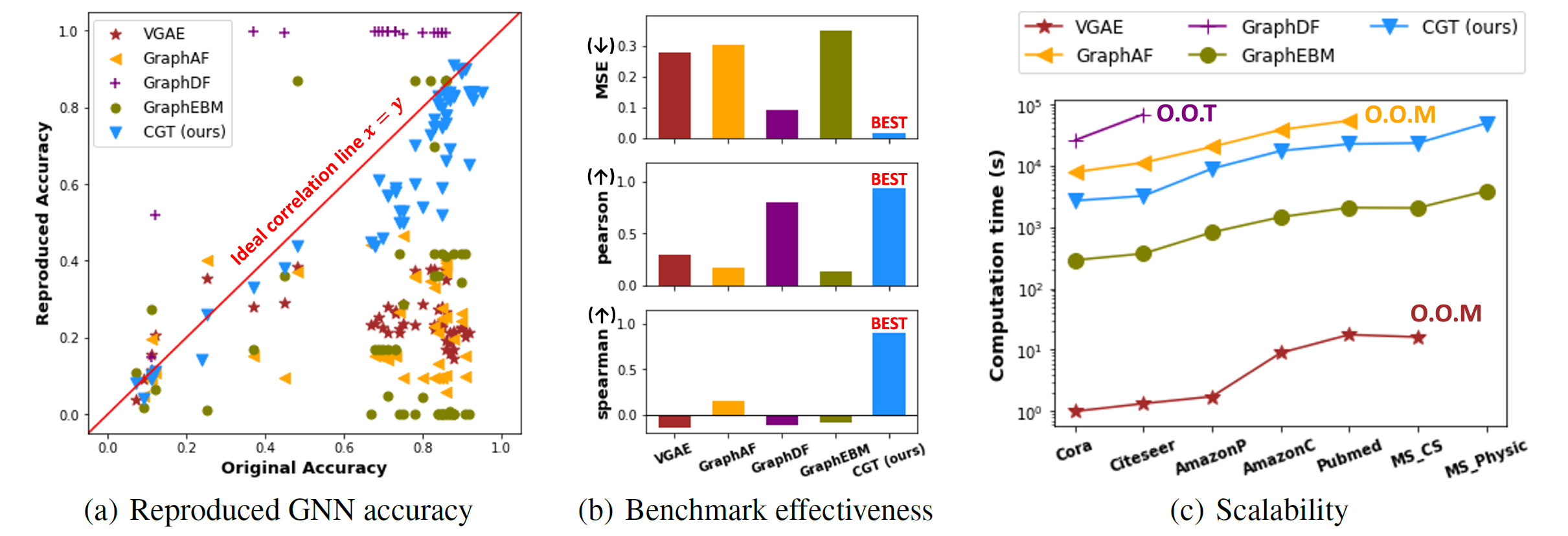
- benchmark effectiveness
- performance ranking on G’ ~ that of G
- scalibility
- linear to a size of graph
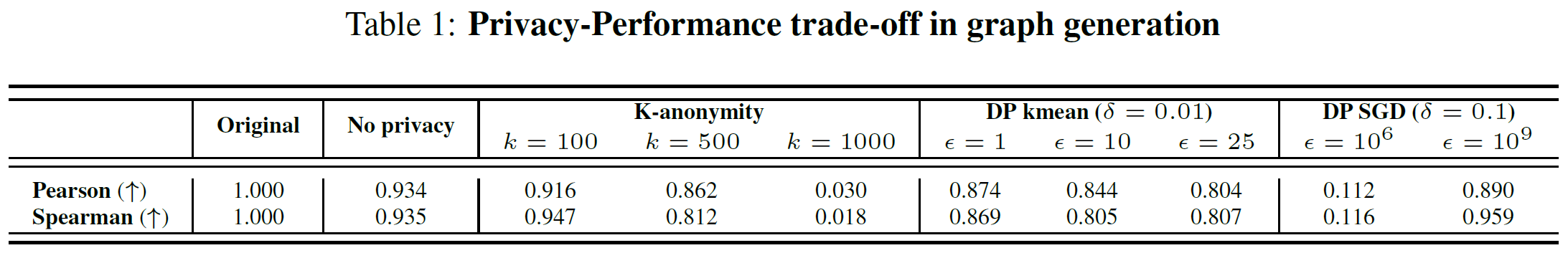
- privacy guarantee
⇒ no study fully addressing the problem
- benchmark effectiveness
- introduce CGT
- reframe: graph generation problem → discrete-value sequence generation problem
- learn distirbution of computation graphs rather than whole graph
- a novel duplicate encoding scheme for computational graphs
- quantize the feature matrix → discrete value sequence
- reframe: graph generation problem → discrete-value sequence generation problem
[2. Related work]
-
traditional graph generative models
⇒ focus on generating graph structures
- general-purpose deep graph generative models
- GAN, VAE, RNN
- evaluation metrics: orbit counts, degree coeffs, clustering coefficients
- doesn’t consider quality of generated node attributes and labels
- molecule graph generative models
[3. From graph generation to sequence generation]
[3.1 Computation graph sampling in GNN training]
- GNNs extract each node v’s egonet (computation graph) instead of operating on the whole graph
- compute embedding of node v on Gv
- problem reduces to:
- given a set of computation graphs {Gv=(Av, Xv, Yv): v\in G} sampled from an original graph, we generate a set of computation graphs {G’v=(A’v, X’v, Y’v)}.
[3.2 Duplicate encoding scheme for computation graphs]
- rules of sampling methods
- #neighbors sampled for each node is limited to keep computation graphs small
- maximum distance from the target node v to sampled nodes is decided by the depth of GNN models
⇒ maximum number of neighbors = neighbor sampling number s
⇒ maximum number of hops from the target node = depth of computation graphs L
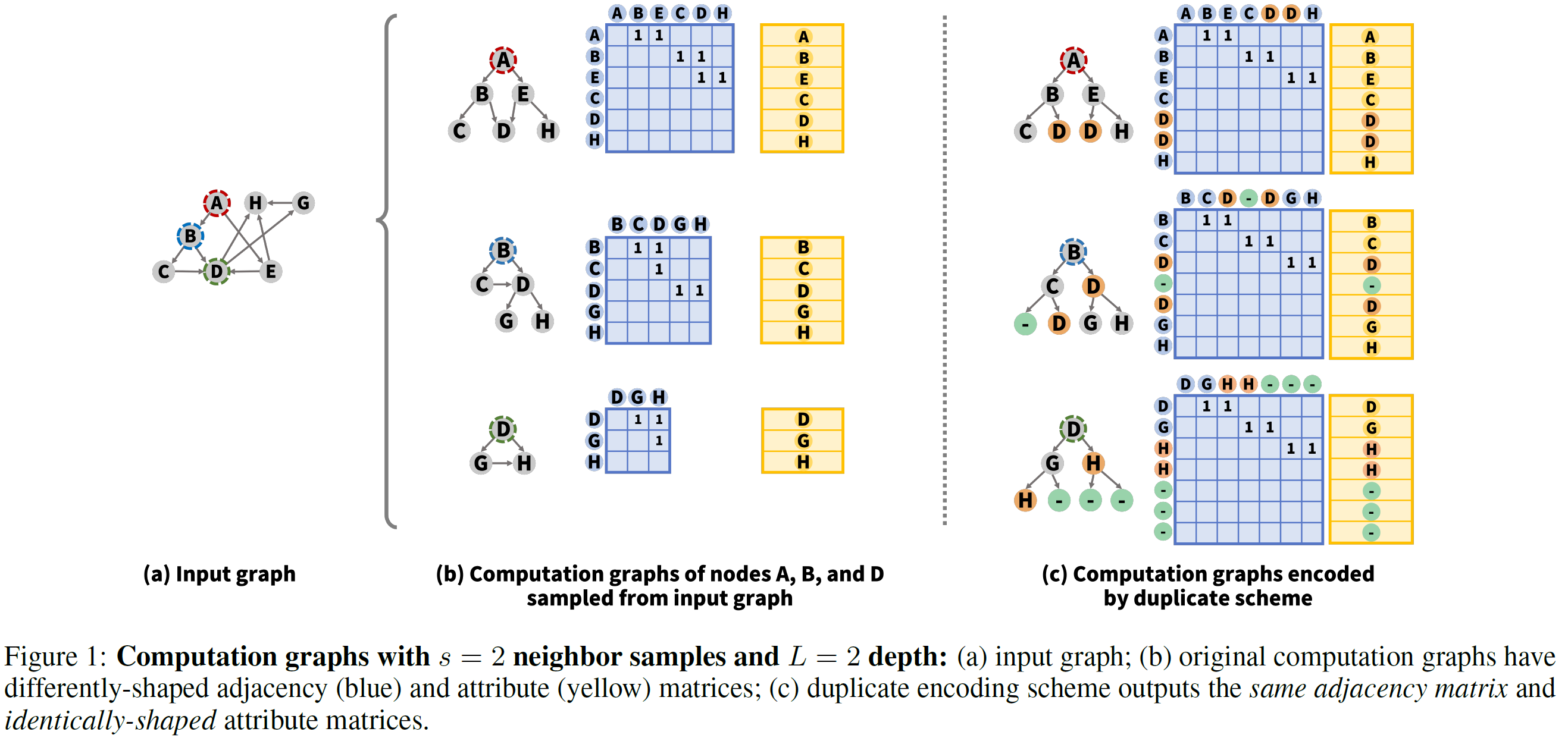
- duplicate encoding scheme
- fixes the structure of all computation graphs ⇒ L-layered s-nary tree structure
- let adjacenccy matrix as a constant
- if deg(v)<s, employ a null node with zero attribute vector, samples it as a padding neighbor
- when a node has a neighbor sampled by another node, duplicate encoding scheme copies the shared neighbor and provides each copy to parent nodes
- have to fix the order of nodes (like, breadth-first ordering)
⇒ given a set of feature matrix-label pairs of duplicated-encoded computation graphs, we generate a set of feature matrix-label pairs
[3.3 Quantization]
- goal: learn distribution of feature matrices of computation graphs
- method: quantize feature vectors into discrete bins
- cluster feature vectors in the original graph using k-means
- map each feature vector to its cluster id
- motivated by
- privacy benefits
- ease of modeling
[3.4. End-to-end framework for a benchmark graph generation problem]
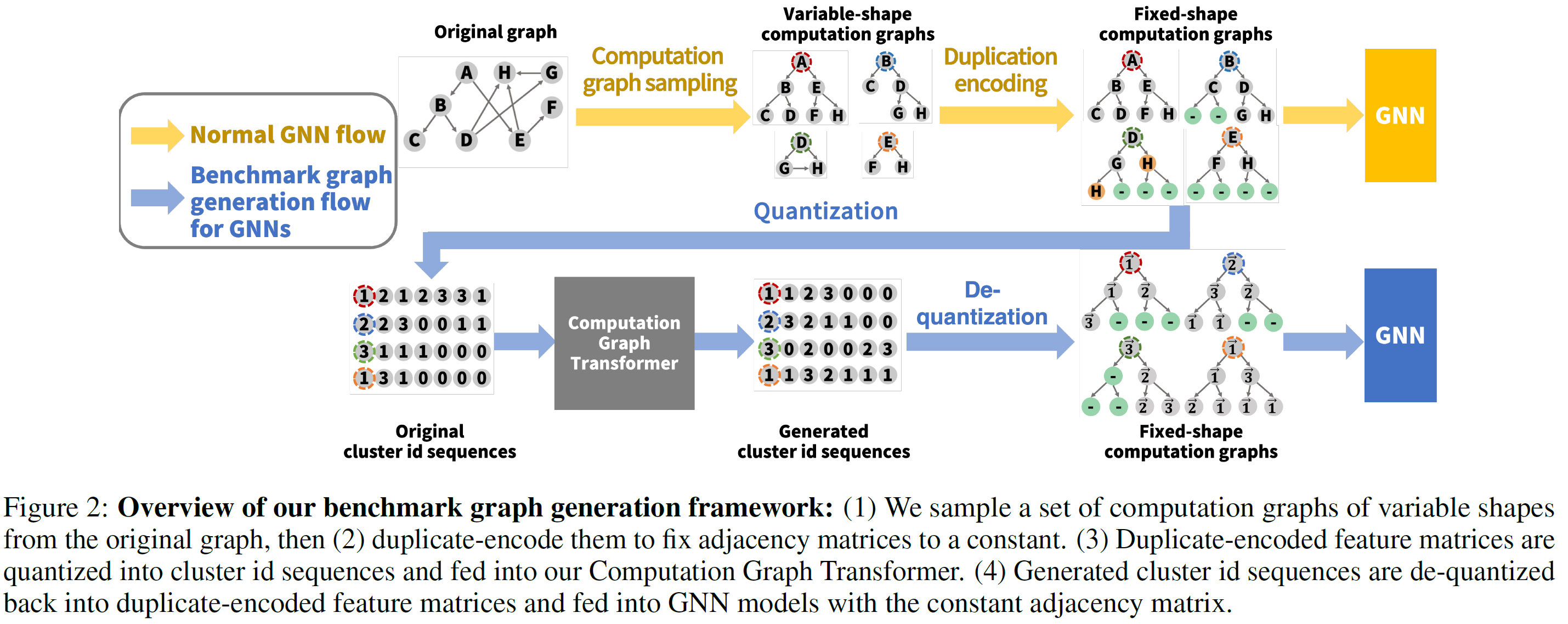
- sample a set of computation graphs from input graph
- encode each computation graph using the duplicate encoding scheme to fix adjacency matrix
- quantize feature vectors to cluster id they belong to
- hand over a set of pairs to new Transformer architecture
- generation process: perform through opposite direction
[4. Model]
- encode computational graph structure → sequence generation process with minmal modification to the Transformer architeucture
[4.1 Computation graph transformer (CGT)]
- extend a two-stream self-attention mechanism, XLNet
-
maximize likelihood
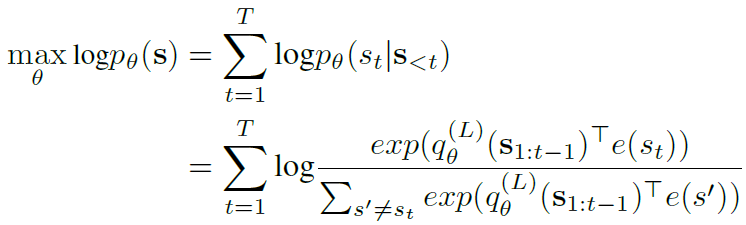
-
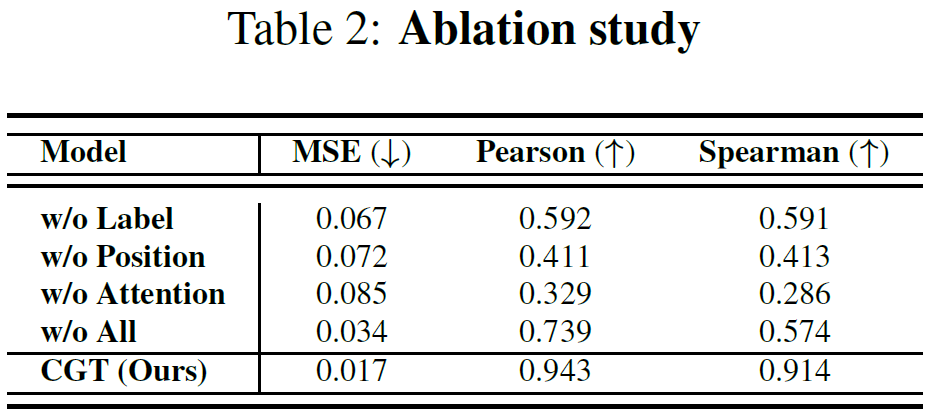
- position embeddings
- sequence (Fig 3(a)) = flattened computational graph (Fig 3(b))
- to encode original computational graph structure, provide differnt position embeddings to different layers in the computation graph
- In figure 3(b), node C, D, F, H located at the 1-st layer have the same position embedding p1
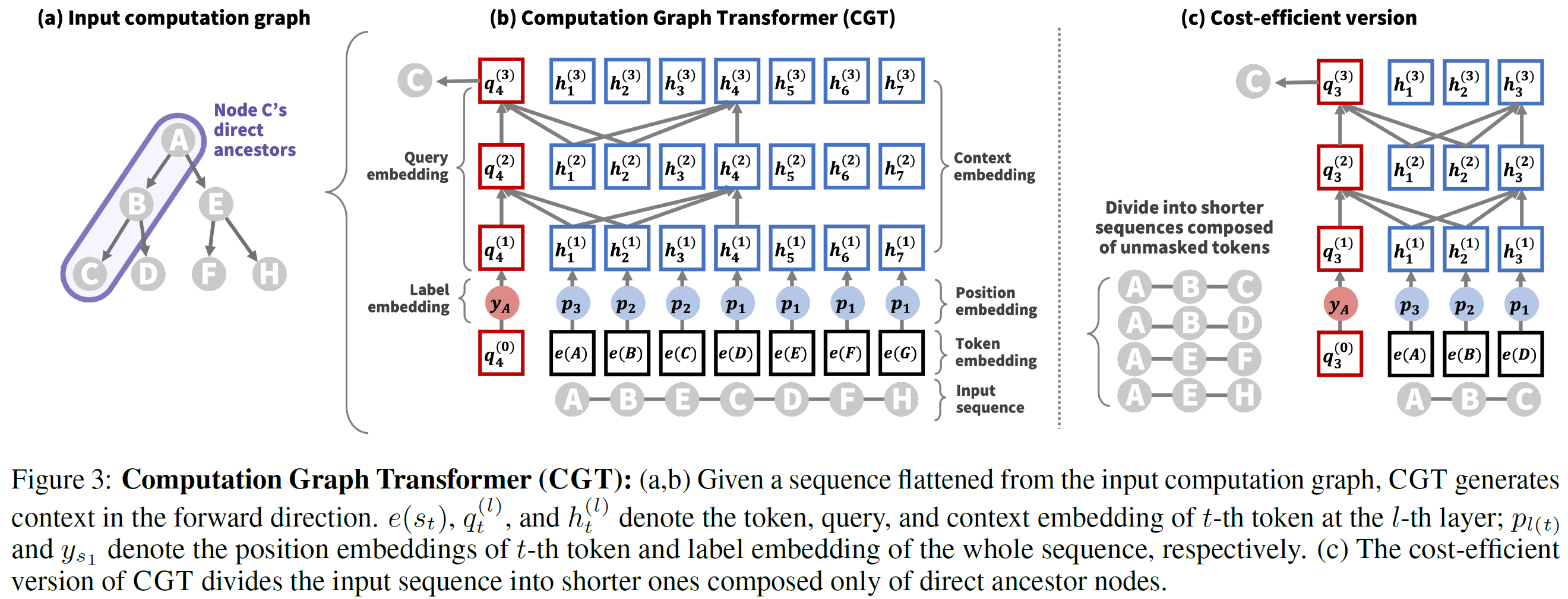
- attention mask
- $q_t^{(l)}$: query embedding
- $h_t^{(l)}$: context embedding
⇒ two above attend to all context embeddings $h^{(l-1)}_{1:t-1}$ before t
- each node in the computation graph is sampled based on its parent node, not directly affected by its sibling nodes
- mask all nodes except direct ancestor nodes in the computation graph
- label conditioning
- distributions of neighboring nodes are not only affected by each node’s feature information but also by its label
- without label information, we can’t learn whether a node has feature-wise homogeneous neighbors or feature-wise heterogeneous neighbors but with the same label
4.2 Theoretical analysis
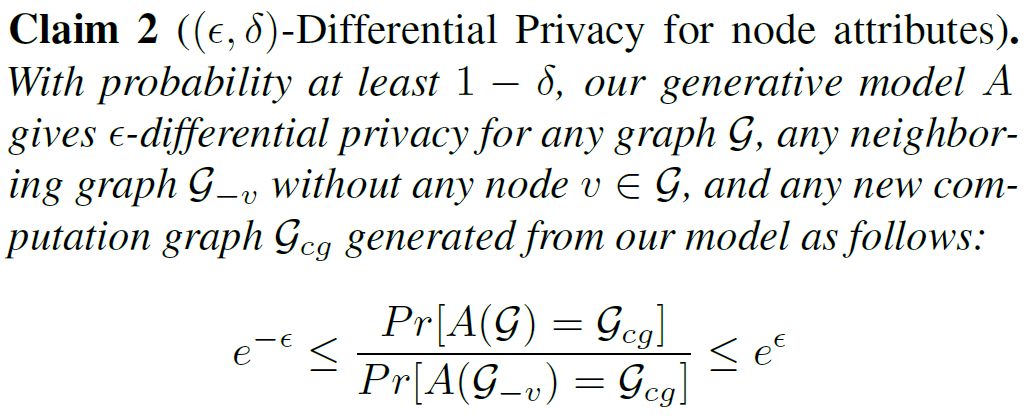
- provide k-anonymity for node attributes and edge distributions (k-means)


[5. Experiments]
[5.1 Experimental setting]
- Baselines: SOTA graph generative models that learn graph structures with node attribute information